Visualizing text embeddings using MotherDuck and marimo
2024/12/11 - 6 min read
BYText embeddings have become a crucial tool in AI/ML applications, allowing us to convert text into numerical vectors that capture semantic meaning. These vectors are often used for semantic search, but in this blog post, we'll explore how to visualize and explore text embeddings interactively using MotherDuck and marimo. Visualizing embeddings helps us understand relationships between different pieces of text, detect patterns, and validate whether our embedding model captures the semantic similarities we expect to see.
For those new to marimo, marimo is a reactive Python and SQL notebook that keeps track of the dependencies between cells and automatically re-runs cells (or marks them stale) when code or UI elements change - similar to how Excel recalculates formulas when you update cell values. This means cells do not get executed from top-to-bottom, but rather, their execution order is determined by the variables, tables, or database created and consumed by each cell. This environment makes it perfect for interactive data exploration.
What We'll Build
By the end of this tutorial, you'll have:
- An interactive visualization of text embeddings in 2D - skip to the demo!
- Automatic clustering of similar texts
- The ability to explore relationships between different pieces of text
- A foundation for building your own text analysis tools
Setting Up Your Environment
You'll need:
- A MotherDuck account, with access to the
embedding()function (a SQL function that converts text to embeddings) marimolocally installed. First, create a new virtual environment with your preferred package manager. Install with:pip install 'marimo[recommended]'or follow the installation instructions.Python >= 3.10
Create a new marimo notebook by running marimo edit embeddings_explorer.py. We'll start by importing the required libraries:
Copy code
# Data manipulation and database connections
import polars as pl
import duckdb
import numba # <- FYI, this module takes a while to load, be patient
import pyarrow
# Visualization
import altair as alt
import marimo as mo
# ML tools for dimensionality reduction and clustering
import umap # For reducing high-dimensional embeddings to 2D
import hdbscan # For clustering similar embeddings
import numpy as np
from sklearn.decomposition import PCA
marimo will automatically ask you to install these dependencies. Choose the package manager you used to install marimo from the dropdown and hit Install. If you would like to start from a reproducible notebook with the same versions, you can download this notebook and run: marimo edit embeddings_explorer.py --sandbox
Connecting to MotherDuck and Loading Sample Data
First, let's connect to MotherDuck. marimo supports both Python and SQL cells, making database operations straightforward:
Copy code
-- This will prompt you to log in and authorize the connection.
ATTACH IF NOT EXISTS 'md:my_db'
This command will open a new browser window to log into MotherDuck. Next, we'll load sample data from the Hacker News Posts dataset. We'll create a table called demo_embedding containing popular posts with a specific keyword. Thanks to marimo's reactivity, any changes to this query will automatically update (or mark stale) any dependent visualizations:
Copy code
CREATE OR REPLACE TABLE my_db.demo_embedding AS
SELECT DISTINCT ON (url) * -- Remove duplicate URLs
FROM 'hf://datasets/julien040/hacker-news-posts/story.parquet'
WHERE contains(title, 'database') -- Filter for posts about databases
AND score > 5 -- Only include popular posts
LIMIT 50000;
Converting Text to Embeddings
Text embeddings are dense vectors that represent the meaning of text. Similar texts will have similar vectors, making them useful for tasks like semantic search and clustering. We'll use MotherDuck's embedding() function to generate these vectors:
Copy code
embeddings = mo.sql(
f"""
SELECT *, embedding(title) as text_embedding
FROM my_db.demo_embedding
LIMIT 1500; -- Limiting for performance in this demo, but you can adjust this
"""
)
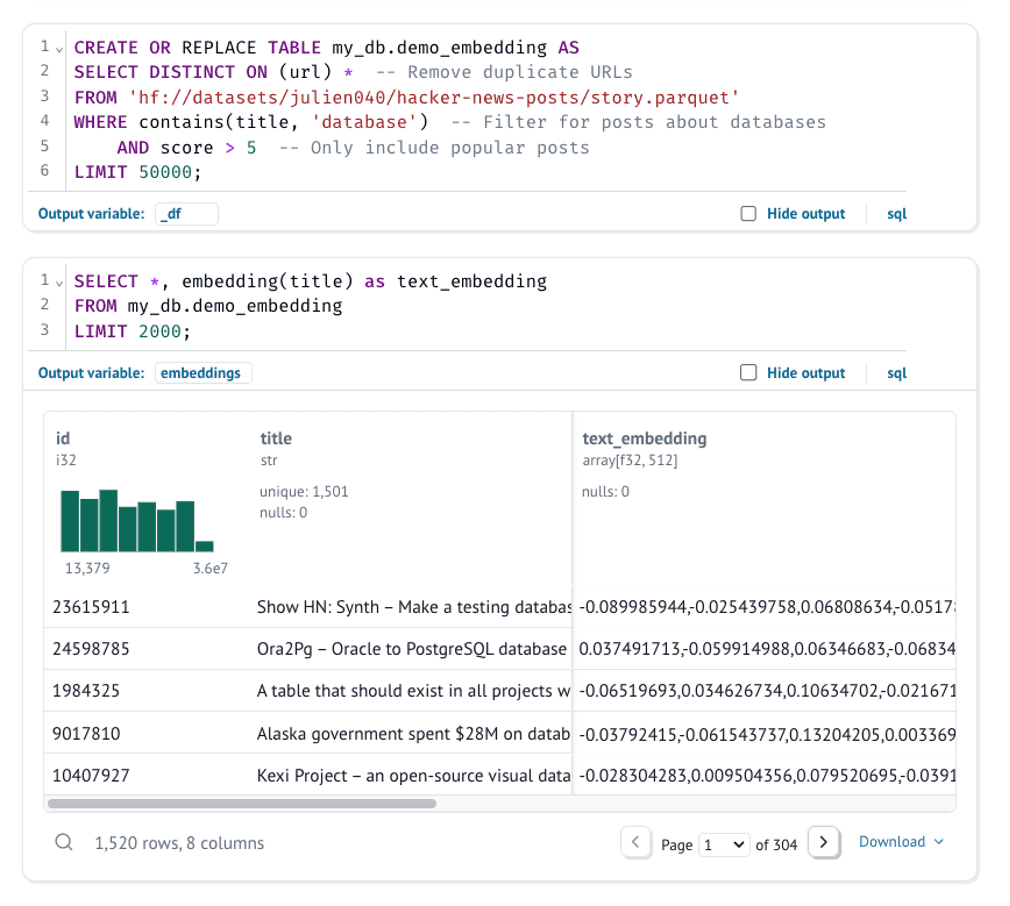
The results are stored in the embeddings variable in Python, which we'll use for clustering and visualization. Each embedding is a high-dimensional vector (in our case, 512 dimensions).
Making Sense of High-Dimensional Data
Text embeddings typically have hundreds of dimensions (512 in our case), making them impossible to visualize directly. We'll use two techniques to make them interpretable:
- Dimensionality Reduction: Convert our 512D vectors into 2D points while preserving relationships between texts
- Clustering: Group similar texts together into clusters
Here are our helper functions:
Copy code
def reduce_dimensions(np_array):
"""
Reduce the dimensions of embeddings to a 2D space.
Here we use the UMAP algorithm. UMAP preserves both local and
global structure of the high-dimensional data.
"""
reducer = umap.UMAP(
n_components=2, # Reduce to 2D for visualization
metric="cosine", # Use cosine similarity for text embeddings
n_neighbors=80, # Higher values = more global structure
min_dist=0.1, # Controls how tightly points cluster
)
return reducer.fit_transform(np_array)
def cluster_points(np_array, min_cluster_size=4, max_cluster_size=50):
"""
Cluster the embeddings.
Here we use the HDBSCAN algorithm. We first reduce dimensionality to 50D with
PCA to speed up clustering, while still preserving most of the important information.
"""
pca = PCA(n_components=50)
np_array = pca.fit_transform(np_array)
hdb = hdbscan.HDBSCAN(
min_samples=3, # Minimum points to form dense region
min_cluster_size=min_cluster_size, # Minimum size of a cluster
max_cluster_size=max_cluster_size, # Maximum size of a cluster
).fit(np_array)
return np.where(hdb.labels_ == -1, "outlier", "cluster_" + hdb.labels_.astype(str))
Processing the Data
Now we'll transform our high-dimensional embeddings into something we can visualize:
Copy code
with mo.status.spinner("Clustering points...") as _s:
embeddings_array = embeddings["text_embedding"].to_numpy()
hdb_labels = cluster_points(embeddings_array)
_s.update("Reducing dimensionality...")
embeddings_2d = reduce_dimensions(embeddings_array)
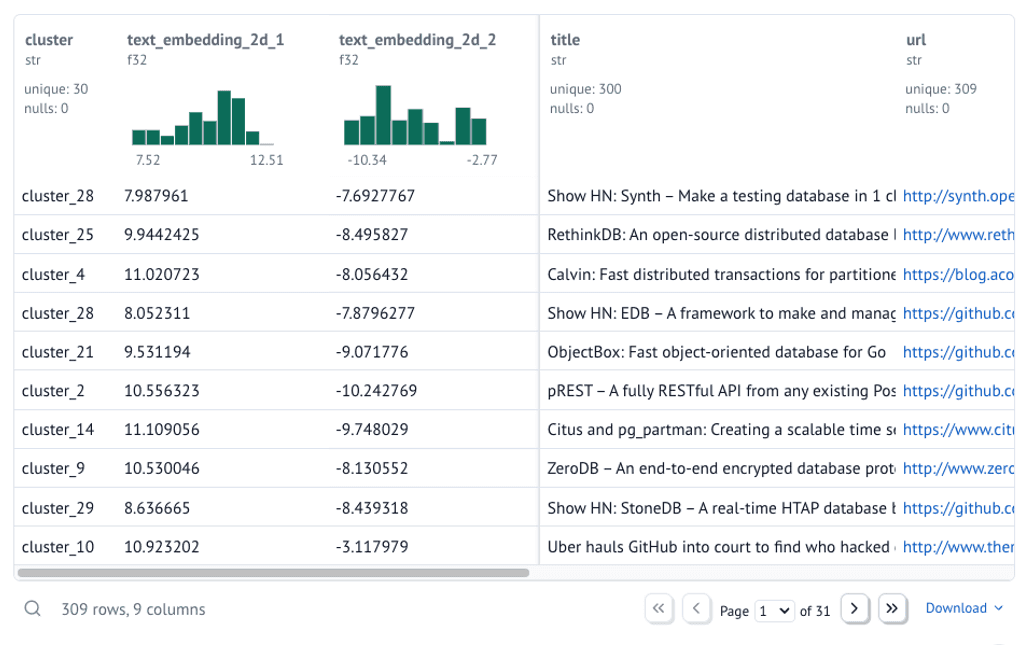
Using polars, we can stitch the 2D embeddings and the cluster labels back on to our original dataframe.
Copy code
data = embeddings.lazy() # Lazy evaluation for performance
data = data.with_columns(
text_embedding_2d_1=embeddings_2d[:, 0],
text_embedding_2d_2=embeddings_2d[:, 1],
cluster=hdb_labels,
)
data = data.unique(subset=["url"], maintain_order=True) # Remove duplicate URLs
data = data.drop(["text_embedding", "id"]) # Drop unused columns
data = data.filter(pl.col("cluster") != "outlier") # Filter out outliers
data = data.collect() # Collect the data
data
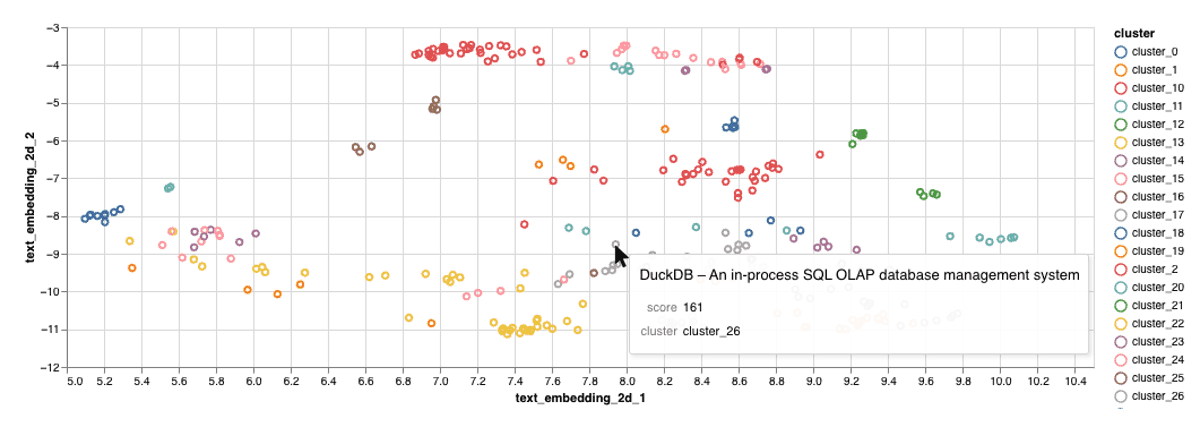
Creating an Interactive Visualization
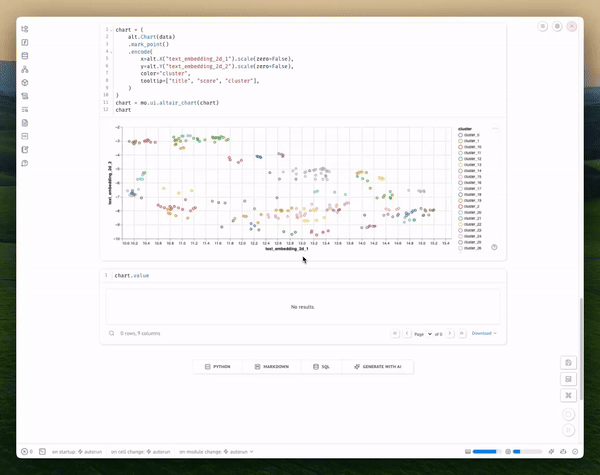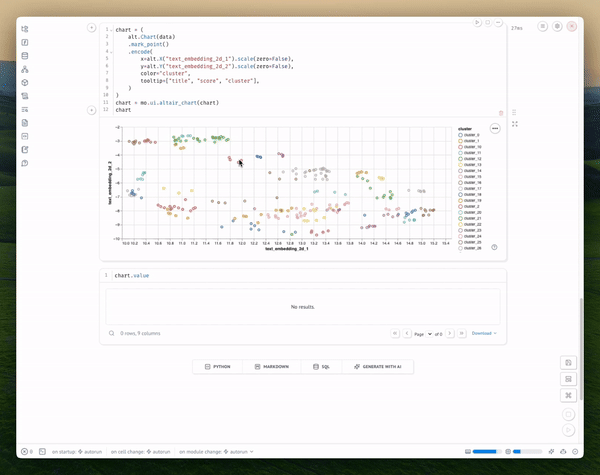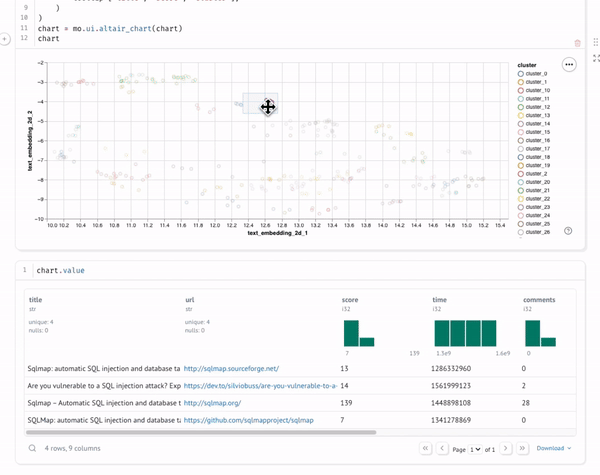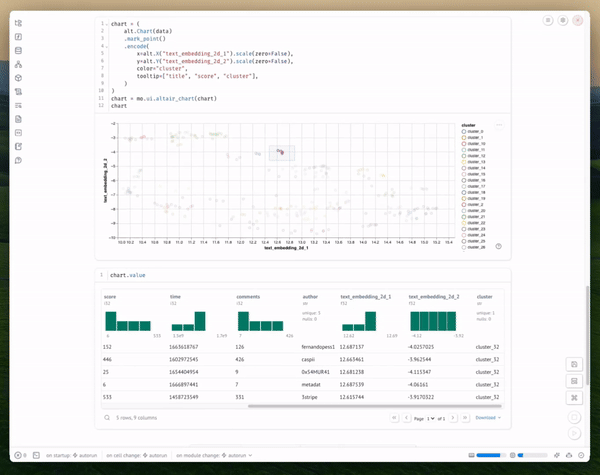
Let's create a scatter plot where:
- Each point represents a text (Hacker News title in our case)
- Similar texts appear closer together
- Colors indicate different clusters of related texts
- You can interact with points to see the underlying text
Copy code
chart = alt.Chart(data).mark_point().encode(
x=alt.X("text_embedding_2d_1").scale(zero=False),
y=alt.Y("text_embedding_2d_2").scale(zero=False),
color="cluster",
tooltip=["title", "score", "cluster"]
)
chart = mo.ui.altair_chart(chart)
chart
And display the chart's selected points:
Copy code
chart.value
Exploring the Results
You can interact with the visualization in several ways:
- Hover over points to see the actual titles
- Look for clusters of related topics
- Identify outliers or unexpected groupings
The visualization shows clusters of semantically similar texts. Each point represents a document, and the proximity between points indicates semantic similarity. Colors represent different clusters identified by our clustering algorithm.
Customizing the Analysis
You can experiment with:
- UMAP parameters:
n_neighbors: Higher values (>100) preserve more global structure, lower values (<20) focus on local relationshipsmin_dist: Controls how tightly points cluster together
- HDBSCAN parameters:
min_cluster_size: Minimum number of points to form a clustermin_samples: Controls noise sensitivity (higher values = more points labeled as noise)
- Different embedding models in MotherDuck:
- Try
embedding(title, model="text-embedding-3-large")for potentially better results
- Try
Next Steps
That's it! You've created an interactive text embedding explorer using MotherDuck and marimo. The full code is available here, as well as an interactive demo deployed as a marimo application.
If you prefer using python directly, its easy as these two commands to get started:
Copy code
pip install marimo marimo edit
Some ideas if you’d like extend this:
- Change the initial dataset
- Choose a different initial keyword or filters
- Add marimo sliders and inputs to make tweaking UMAP and HDBSCAN even easier
- Implement semantic search functionality to highlight related points
PREVIOUS POSTS
2024/12/04 - Jordan Tigani
Introducing Read Scaling
Read Scaling is now in preview! Read Scaling improves DuckDB SQL query performance by scaling out to multiple DuckDB instances, known as Read Scaling replicas. It is useful to speed up BI dashboards and data apps significantly.
2024/12/09 - Sheila Sitaram
The Serverless Backend for Analytics: Introducing MotherDuck’s Native Integration on Vercel Marketplace
MotherDuck's native integration is now available on Vercel Marketplace. Developers can finally streamline their application maintenance overhead when building embedded analytics components and data apps. Start building with templates and a demo app!

